This is for my various creationist readers who keep (mistakenly) proclaiming that multi-protein systems can’t evolve. I’ve shown why this isn’t true, but it’s time for another one. Again, this paper directly refutes those claims and shows that evolution CAN result in multi-protein systems.
Two of the authors (but not the lead author) are people that are familiar to those in the community; Sean M. Carrol and Joseph W. Thorton. The paper’s lead author is Jamie T. Bridgham and it is titled Evolution of Hormone-Receptor Complexity by Molecular Exploitation.*
So, the abstract opens with the same complain that creationists have.
According to Darwinian theory, complexity evolves by a stepwise process of elaboration and optimization under natural selection. Biological systems composed of tightly integrated parts seem to challenge this view, because it is not obvious how any element’s function can be selected for unless the partners with which it interacts are already present.
So, unlike creationists, these three sat down and did some science and came up with an answer to the question. I’d like to point out that this research paper was published in 2006. Since it doesn’t say “We discover how tightly integrated biological system evolved”, then it’s easy to see how creationists miss this. Whenever you hear this argument, give them a link to this paper and then enjoy the change in subject.
A particular steroid hormone (aldosterone) and it’s specific receptor partner (MR) were examined. MR and the related GR were formed from a gene duplication way back in the ancestor of vertebrates. GR is activated by cortisol (a stress hormone) to regulate metabolism and inflammation. MR is activated by aldosterone and controls some aspects of homeostasis.
While MR can be activated by cortisol, it doesn’t happen often because of some other biochemical effects. The question is, how did MR develop such a tight affinity for aldosterone? To quote the authors (and every creationist)
If the hormone is not yet present, how can selection drive the receptor’s affinity for it? Conversely, without the receptor, what selection pressure could guide the evolution of the ligand?
The authors looked for the ancestral form of the MR/GR common ancestor. The fact that this can be done in a statistically robust fashion is proof enough of common ancestry. They identified a corticoid receptor in jawless fishes (the most primitive** organism to not have GR and MR) and both GR and MR in skates. This data helped determine that the GR/MR split occurred more than 450 million years ago.
They recreated this primitive protein that they called the ancestral corticoid receptor (AncCR). They found it was very sensitive to aldosterone, but also activated by low doses of cortisol. Let me add that this also isn’t just a flight of fancy, but highly statistically robust. If you get to arguing with a creationist and they aren’t talking about statistical data, then they don’t have any idea how this work is done and therefore are not in any position to criticize it.
Interestingly, the authors thought that they were looking for how MR became associated with alodsterone, but what they found was that aldosterone affinity was the original considition and corticoid affinitiy came later. This is doubly interesting because only tetrapods produce aldosterone. So, the ability to bind with aldosterone appeared much sooner than aldosterone itself did.
So, that sort of punches a hole in the problem right there. It is perfectly plausible that this happened with other protein systems. Just because a protein does a certain thing doesn’t mean that it only does that thing.
The authors then started looking at how to get from the ancestral protein to the modern protein. The GR obviously lost the ability to bind with aldosterone. By mutating the ancestral form, they found a version that increased it’s inability to bind with aldosterone by three orders of magnitude and was still able to moderately bind with cortisol. This occurred due to two changes in the protein.
An analysis of the human GR shows that it is these two changes that exist in our own system. So, what the researchers did was create an ancestral protein, try to mutate it so it had the same effect as the modern protein, then compared that to the actual modern protein.
This is a huge vindication for evolution. Not only does it work, but it has predictive power.
Our findings demonstrate that the MR-aldosterone partnership evolved in a stepwise fashion consistent with Darwinian theory, but the functions being selected for changed over time. AncCR_s sensitivity to aldosterone was present before the hormone, a by-product of selective constraints on the receptor for activation by its native ligand. AncCR and its descendant genes were structurally preadapted for activation by aldosterone when that hormone evolved millions of years later. After the duplication that produced GR and MR, only two substitutions in the GR lineage were required to yield two receptors with
distinct hormone-response profiles. The evolution of an MR that could be independently regulated by aldosterone enabled a more specific endocrine response, because it allowed electrolyte homeostasis to be controlled without also triggering the GR stress response, and vice versa.
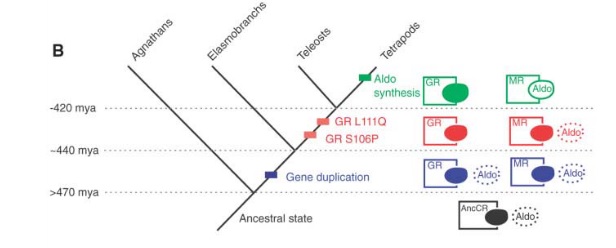
This picture shows the results very clearly.
The ancestral state (AncCR; in black) could slightly bind with aldosterone. There was a gen duplication event somewhere in the 470-440 million years ago range. Both genes (in blue) built proteins that could bind with aldosterone. Then, somewhere between 440-420 million years ago, the GR version lost the ability to bind with aldosterone.
Of course, to this point, aldosterone didn’t even exist. The earliest tetrapods existed about 395 million years ago, so the appearance of aldosterone occurred somewhen between then and 420 million years ago.
While the MR/aldosterone system may look complex and one wonders how could the protein evolve before it could do anything and how could the receptor evolve before there was anything to receive, the conclusion is radically more simple.
The receptor had the ability long before the protein it binds to existed. And this is because, proteins (unlike the vast majority of designed things that we know of) can do multiple things like binding with multiple targets.
They didn’t have to evolve together. This research shows, quite clearly, that the one evolved tens of millions of years before the other.
_________________________________
*Bridgham, J., Carroll, S. & Thornton, J. Evolution of hormone-receptor complexity by molecular exploitation. Science (New York, N.Y.) 312, 97–101 (2006).
** I use primitive in the evolutionary sense.