I’m an odd person. When I see some clown on the internet make a claim and the very next issue of Science has a paper that utterly demolishes that claim, I get all excited.
After the debates, I’ve had several conversations with creationists who make the claim that mutations can’t do something. Mutations can’t create new structures. Mutations change the gene and make it non-functional. Mutations can’t create new information. Blah blah blah. The simple truth is that they can. The paper in question is by Camille Sayou et. al. (It took 14 people to do this research.)
As always, if something I say is unclear or doesn’t make sense, please let me know.
Most genes in any organism actually don’t make proteins that build the organism or make muscles or whatever else. Most genes make proteins that regulate other genes, turning them on or off or speeding up or slowing down their transcription. This changes the amount of that protein in the organism and will have some kind of effect.
These regulatory proteins are called transcription factors. They are important drivers in evolution. A mutation in a transcription factor can result in a longer than normal body or more of a particular blood protein. This may provide an advantage or may kill the organism before it gets past the blastula stage.
People often wonder how mutations can cause extra limbs (for example) to form. Well, it’s not a change in the muscle proteins and the bone proteins and the skin proteins. It’s one change in the regulatory protein that controls all the others. Small changes, in the right spot can have massive effects on the final organism.
These leads to the question of how can these genes evolve at all? If they are so fraught with danger, changing those regulatory proteins should almost universally be bad right? Well… pretty much. Yeah. But (and there’s always a ‘but’ isn’t there?), many of these genes have multiple copies in each organism. This often results from DNA copy errors or chromosome separation errors. Regardless of how it happens, the organism now has two copies of the gene. One of which fulfills the essential function of the gene and one is free to mutate without harming the organism.
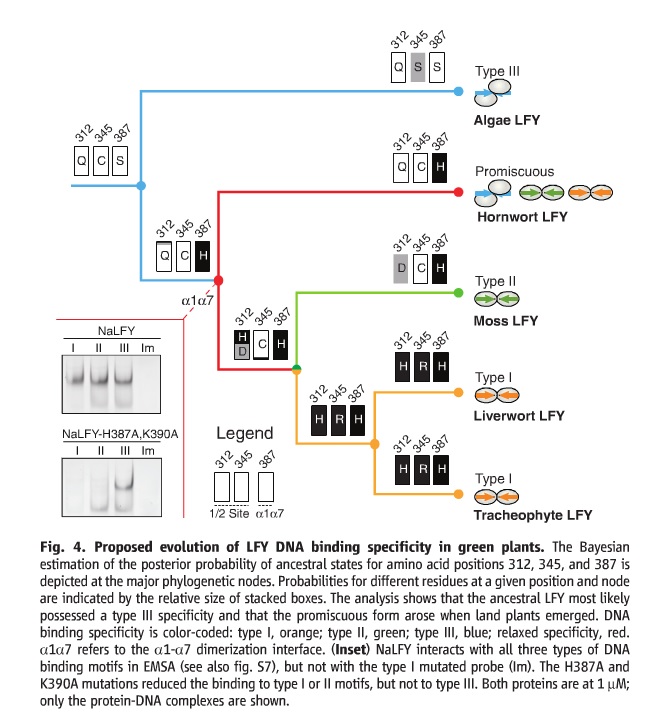
The gene Dr. Sayou (and the others) looked at is called LEAFY. This gene codes for a transcription factor that is a major regulator of flower development and cell division in land plants. What is interesting though, is that the vast majority of species of plants have only a single copy of LEAFY. What’s even more interesting is that there are three distinct types of LEAFY. I will refer to these as Type 1, Type 2, and Type 3.
Most land plants are Type 1. This includes flowering plants, cone bearing plants (which happens to have a parlogue called NEEDLY), the ferns, and the liverworts. Type 2 includes the mosses. Type 3 includes the hornworts and, curiously, algae. Yep, this gene goes way back.
They are obviously the same gene with only minor mutational differences. In fact all the differences occur because of changes at 3 different points. So, the question is, how do we get these wildly different versions of a gene that is very specific and very conserved?
The answer is, biochemistry. The authors analyzed the protein that LEAFY makes in many plants and analyzed the results.
This cladogram is the result. The three boxes labeled “312”, “345”, and “387” t each junction represent the mutations in the transcription factor at those numbered positions (it’s a big protein). By analyzing even algal samples, they were able to determine the ancestral version (to a high confidence). That ancestral version is on the upper left, QCS.
The algae version is most like the ancestral version, which is to be expected. The split at QCH is what is interesting to us, because that’s where the Type 2 and Type 3 versions come from. But if this is so particular, how can it change?
Because that one mutation, changing position 387 from an S amino acid to a H amino acid (yes, I could go look them up, no I don’t care right now) results in a version of the transcription factor that the authors call “Promiscuous”. That promiscuous version of the transcription factor is capable of performing the functions of Type 1 AND type 2 AND Type 3 LEAFY.
It’s transitional!
But, if it’s so cool that it can function as all three versions, why did the other version appear (Type 2 and Type 1)? Because it’s not quite as efficient at all of the functions as the versions on the right are. It’s like having a phone that’s also a camera that’s also a tablet computer. That’s very cool, but try to whip out your phone and take that super awesome snapshot. You’ll probably miss because the phone just isn’t as efficient as a point and shoot camera.
In fact, the authors specifically mention that they missed noticing that the promiscuous version could even act as Type 1 or TYpe 2 because it was less efficient at them.
So there you have it. Not only can mutations change genes so we get new effects on the organisms, but in at least one case (LEAFY) we know that mutations in important transcription factors don’t even have to disrupt the organisms that much.
This is very, very cool research and puts another nail in the “mutations can’t” myth.
_______________________________
C. Sayou et al., A Promiscuous Intermediate Underlies the Evolution of LEAFY DNA Binding Specificity.Science (New York, N.Y.) (2014), doi:10.1126/science.1247660