Yesterday (April 17th), Nature published the results of the sequencing of the coelacanth genome (this is the open access paper).

from http://en.wikipedia.org/wiki/File:Coelacanth1.JPG
The above is a coelacanth (I just like saying it, it’s a neat word). The scientific name is Latimeria chalumnae named after Marjorie Courtenay-Latimer, the curator of a small natural history museum in South Africa. She discovered the creature in 1938, after picking through the catch from local fisherman.
At the time, it was though that the coelacanth’s had gone extinct 70 million years prior.
At this point people start talking about ‘living fossils’ and crap like that. No, no, no. Evolution happens. It’s a fact. While the coelacanth’s of today are morphologically similar, they are as distinct from their ancestors of 70 millions years ago as birds are distinct from dinosaurs.
The paper confirms that these animals are not exactly the same as the fossil ones. Of course, that being said, it is very interesting that the rate of mutation and genetic change in coelacanth’s seem to be lower than most anything else. The author’s suspect that because they are such deep water fishes (i.e. a very stable environment) and little or no predation or competition, that adaptation is not the massive advantage it would be in terrestrial species (for example).
Biologists have thought since 1938 that the coelacanth might be the closest living relative of terrestrial species. They are members of Sarcopterygii, lobe-finned fishes and all tetrapods (i.e. all vertebrate land animals) along with lungfish.
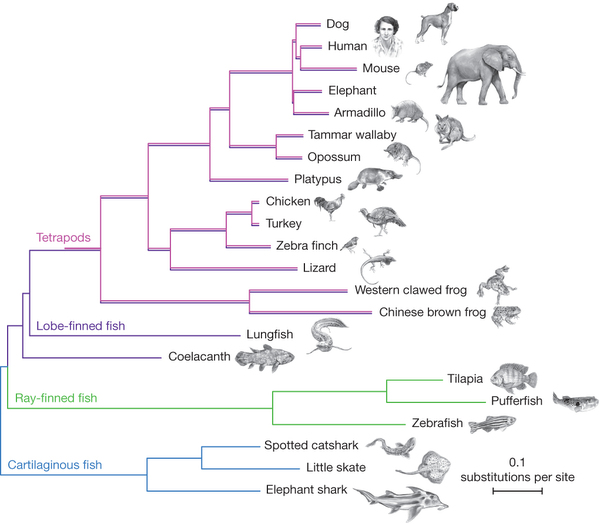
This cladogram was developed from 251 genes in all species. The results are statistically at 100% (except for the Armadillo/Elephant positions at 45%). This cladogram is interesting in that the length of the lines represent (effectively) the rate of evolution (in substitutions per site).
Statistically, it is clear that lungfish are the nearest living relative to terrestrial vertebrates. And that coelacanths are evolving slowly compared to everything else.
The report describes some really interesting things about coelacanth’s.
Based on the coelacanth, it is now inferred that of the 50 or so genes that are present in fishes and not present in terrestrial vertebrates, 4 were definitely lost lost in the tetrapod lineage. The coelacanth has them, other fishes have them, but tetrapods don’t.
This next one is very interesting. There are sixcis-regulatory sequences in tetrapod genes (for example, a mouse) that are involved in digit formation (this is pretty simplified, so work with me). of those six, three are restricted to tetrapods. One of them, called “island 1”, is also present in the coelacanth, but not present in other fishes.
What’s really interesting is that the coelacanth version was able to promote expression in the mouse. The conclusion is that island 1 was involved in the development of lobe-fins in fishes and then co-opted into a digit (finger) maker in tetrapods.
Similar research shows how urea (pee) as a method to get rid of nitrogenous waste was a key innovation to moving from water to land.
Coelacanth’s (at least the modern ones) give birth to live offspring and have large vascularised eggs. It was determined that the same genes from the Hox family are in coelacanths, some sharks (but it is a pseudogene), and all tetrapods. Indeed, it is so highly conserved that the gene is 99% the same in all mammals.
That gene from coelacanths was used in several transgenic experiments and showed high expression just outside of chick embryos.
Finally, and this one is for you Michael Behe, The coelacanth lacks Immunoglobulin-M (IgM), a class of antibodies that is considered vital for adaptive immunity and is been found in every vertebrate to date.
The researchers did find two IgW genes, which are also immunoglobulin genes, but only found in lungfish and cartilaginous fishes (sharks and rays). These IgW genes have been lost in ray-finned fishes and tetrapods.
The question is, how does the coelacanth respond to new pathogens and do the IgW genes (somehow) play the same role as IgM? Obviously the coelacanth survives quite well, surviving a number of major extinction events. So Behe, perhaps you would care to explain how ID, which predicts that this can’t happen, explains how it happened.
This is some fascinating research. These are very cool fish. But they are not living fossils. They are just as modern as humans are. They are just as different from their ancestors as humans are from our ancestors… except their ancestors living 70 million years ago instead of 7 million years ago.
